Moore, CM;
Dhillon, J;
Flynn, R;
Gizynski, K;
Adams, C;
Morgan, G;
McGurk, D;
Boada, E;
Shabestary, S;
Peat, J;
et al.
Moore, CM; Dhillon, J; Flynn, R; Gizynski, K; Adams, C; Morgan, G; McGurk, D; Boada, E; Shabestary, S; Peat, J; O'Halloran, J; Stoker, NG; Butcher, PD; Murton, H
(2023)
A Novel Microfluidic Dielectrophoresis Technology to Enable Rapid Diagnosis of Mycobacteria tuberculosis in Clinical Samples.
The Journal of Molecular Diagnostics, 25 (7).
pp. 513-523.
ISSN 1525-1578
https://doi.org/10.1016/j.jmoldx.2023.04.005
SGUL Authors: Butcher, Philip David
|
PDF
Published Version
Available under License Creative Commons Attribution. Download (991kB) | Preview |
|
![[img]](https://openaccess.sgul.ac.uk/115461/10.hassmallThumbnailVersion/1-s2.0-S1525157823000983-figs1_lrg.jpg)
|
Image (JPEG) (Supplemental Figure S1)
Published Version
Available under License Creative Commons Attribution. Download (221kB) | Preview |
|
|
Video (MP4) (Supplemental Video S1)
Accepted Version
Available under License Creative Commons Attribution. Download (10MB) |
||
|
Microsoft Word (.docx)
Accepted Version
Available under License Creative Commons Attribution. Download (124kB) |
||
|
Microsoft PowerPoint (Figures)
Accepted Version
Available under License Creative Commons Attribution. Download (3MB) |
Abstract
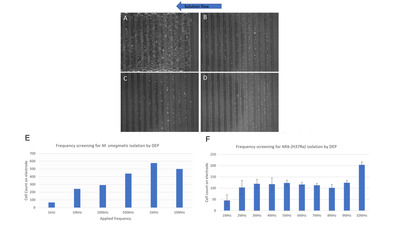
To achieve the global efforts to end tuberculosis, affordable diagnostics suitable for true point-of-care implementation are required to reach the missing millions. In addition, diagnostics with increased sensitivity and expanded drug susceptibility testing are needed to address drug resistance and to diagnose low-bacterial burden cases. The laboratory-on-a-chip technology described herein used dielectrophoresis to selectively isolate Mycobacterium tuberculosis from sputum samples, purifying the bacterial population ahead of molecular confirmation by multiplex real-time quantitative PCR. After optimization using a panel of 50 characterized sputum samples, the performance of the prototype was assessed against the current gold standards, screening 100 blinded sputum samples using characterized and biobanked sputum provided by Foundation for Innovative New Diagnostics. Concordance with culture diagnosis was 100% for smear-negative samples and 87% for smear-positive samples. Of the smear-positive samples, the high burden sample concordance was 100%. Samples were diagnosed on the basis of visual assessment of the dielectrophoresis array and by multiplex real-time quantitative PCR assay. The results described herein demonstrate the potential of the CAPTURE-XT technology to provide a powerful sample preparation tool that could function as a front-end platform for molecular detection. This versatile tool could equally be applied as a visual detection diagnostic, potentially associated with bacterial identification for low-cost screening or coupled with an expanded PCR assay for genotypic drug susceptibility testing.
| Item Type: | Article | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Additional Information: | Copyright © 2023 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0). | ||||||||
| Keywords: | 1108 Medical Microbiology, Pathology | ||||||||
| SGUL Research Institute / Research Centre: | Academic Structure > Infection and Immunity Research Institute (INII) | ||||||||
| Journal or Publication Title: | The Journal of Molecular Diagnostics | ||||||||
| ISSN: | 1525-1578 | ||||||||
| Dates: |
|
||||||||
| Publisher License: | Creative Commons: Attribution 4.0 | ||||||||
| Projects: |
|
||||||||
| URI: | https://openaccess.sgul.ac.uk/id/eprint/115461 | ||||||||
| Publisher's version: | https://doi.org/10.1016/j.jmoldx.2023.04.005 |
Statistics
Actions (login required)
 |
Edit Item |